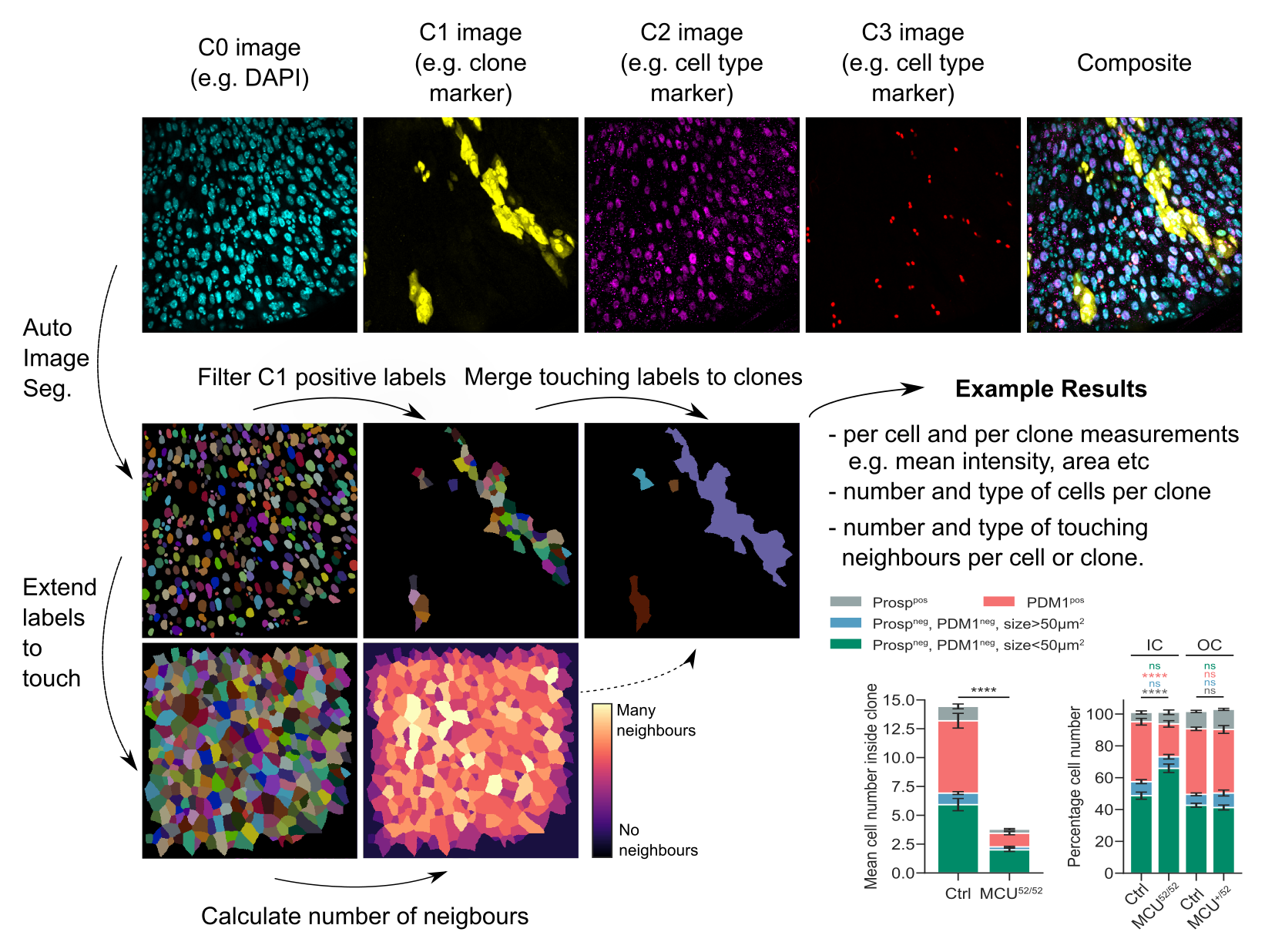
clonedetective is a package for analysing fluorescent imaging data from cell lineage experiments (e.g. FLP-out, MARCM or Cre-lox clones).
Outputted quantifications include:
- counts of each cell type per “clone”
- spatial metrics e.g. number of nearest neighbors of each cell per clone
- cell and clone properties e.g. area, mean intensity etc.
If clones label genetic mutations, these metrics can be useful in addressing biological questions such as:
- does my gene of interest regulate clone size (i.e. cell proliferation) or clone composition (i.e. cell differentiation)?
- does the local cell neighbourhood (e.g. number and type of neighbours) non-autonomously impact cell proliferation or differentiation?
- does my gene of interest regulate the expression of other (immunostained) proteins?
clonedetective is constructed using many amazing python libraries, including scikit-image, numpy, Xarray, pandas, numba, Dask, Dask-image and pyclesperanto-prototype.
Many functions are lazy-loaded and parallelized using Dask, enabling clonedetective to scale to large multi-dimensional datasets that do not fit in RAM.
It is recommended to install clonedetective into a virtual environment e.g. using conda.
If you have anaconda or miniconda installed, you can create a virtual environment using the following command. It is often helpful to install something into an empty environment, in this case we install scipy:
conda create -n myenv scipy
We next activate our environment:
conda activate myenv
You can then install clonedetective into this environment via pip:
pip install clonedetective
How to use
Please see the tutorials:
1) Example walkthrough
2) Downstream Analysis using Python
3) Downstream Analysis using R
4) Generating nuclei segmentation using StarDist
In progress, more to come!